Secretory Capacity and Specificity
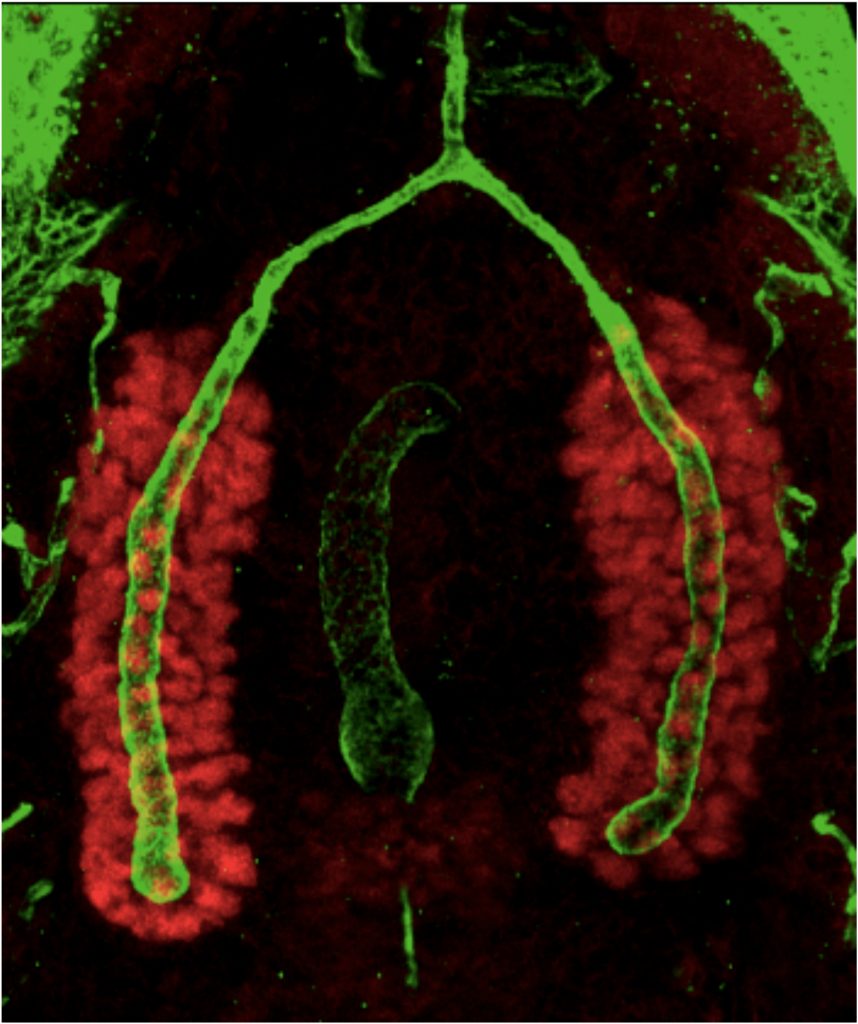
 The Andrew Lab has discovered that the CrebA/Creb3-like bZIP transcription factors are direct and major regulators of secretory capacity. Drosophila CrebA directly activates high-level expression of Secretory Pathway Component Genes (SPCGs) through a site we found to be conserved among the enhancers of CrebA-dependent genes. Moreover, ectopic expression of CrebA in multiple tissues is sufficient to activate high-level expression of every SPCG tested. Microarray analysis indicates that CrebA is required for full expression of ~400 genes including almost 200 already implicated in secretion. The secretory target genes include general machinery required for secretion in all cells as well as cell-type specific secreted cargo, such as the cuticle proteins and mucins. Phenotypic characterization of CrebA mutant SGs reveals a range of expected secretory defects, including reduced luminal secretory content and a decrease in the size and number of apical secretory vesicles, as well as unexpected changes in organelle distribution. In collaboration with Peter O’Hare’s group, we demonstrated that active forms of all five human CrebA orthologues: Creb3/Luman, Creb3L1/Oasis, Creb3L2/BBF2H7, Creb3L3/CrebH and Creb3L4/Creb4 can activate the Drosophila SPCGs when expressed in embryos. Active Creb3L1 also induces expression of multiple components of the secretory pathway when expressed in HeLa cells, a non-secretory cell type. These studies reveal an ancient and highly-conserved role for the CrebA/Creb3-like bZIP transcription factors in boosting secretory capacity. Current efforts are directed toward learning how CrebA interacts with target gene DNA in vivo (in collaboration with the research team of Matthew Slattery at the University of Minnesota, Duluth) and characterizing novel secretory pathway components identified based on their CrebA regulation.
The Andrew Lab has discovered that the CrebA/Creb3-like bZIP transcription factors are direct and major regulators of secretory capacity. Drosophila CrebA directly activates high-level expression of Secretory Pathway Component Genes (SPCGs) through a site we found to be conserved among the enhancers of CrebA-dependent genes. Moreover, ectopic expression of CrebA in multiple tissues is sufficient to activate high-level expression of every SPCG tested. Microarray analysis indicates that CrebA is required for full expression of ~400 genes including almost 200 already implicated in secretion. The secretory target genes include general machinery required for secretion in all cells as well as cell-type specific secreted cargo, such as the cuticle proteins and mucins. Phenotypic characterization of CrebA mutant SGs reveals a range of expected secretory defects, including reduced luminal secretory content and a decrease in the size and number of apical secretory vesicles, as well as unexpected changes in organelle distribution. In collaboration with Peter O’Hare’s group, we demonstrated that active forms of all five human CrebA orthologues: Creb3/Luman, Creb3L1/Oasis, Creb3L2/BBF2H7, Creb3L3/CrebH and Creb3L4/Creb4 can activate the Drosophila SPCGs when expressed in embryos. Active Creb3L1 also induces expression of multiple components of the secretory pathway when expressed in HeLa cells, a non-secretory cell type. These studies reveal an ancient and highly-conserved role for the CrebA/Creb3-like bZIP transcription factors in boosting secretory capacity. Current efforts are directed toward learning how CrebA interacts with target gene DNA in vivo (in collaboration with the research team of Matthew Slattery at the University of Minnesota, Duluth) and characterizing novel secretory pathway components identified based on their CrebA regulation.
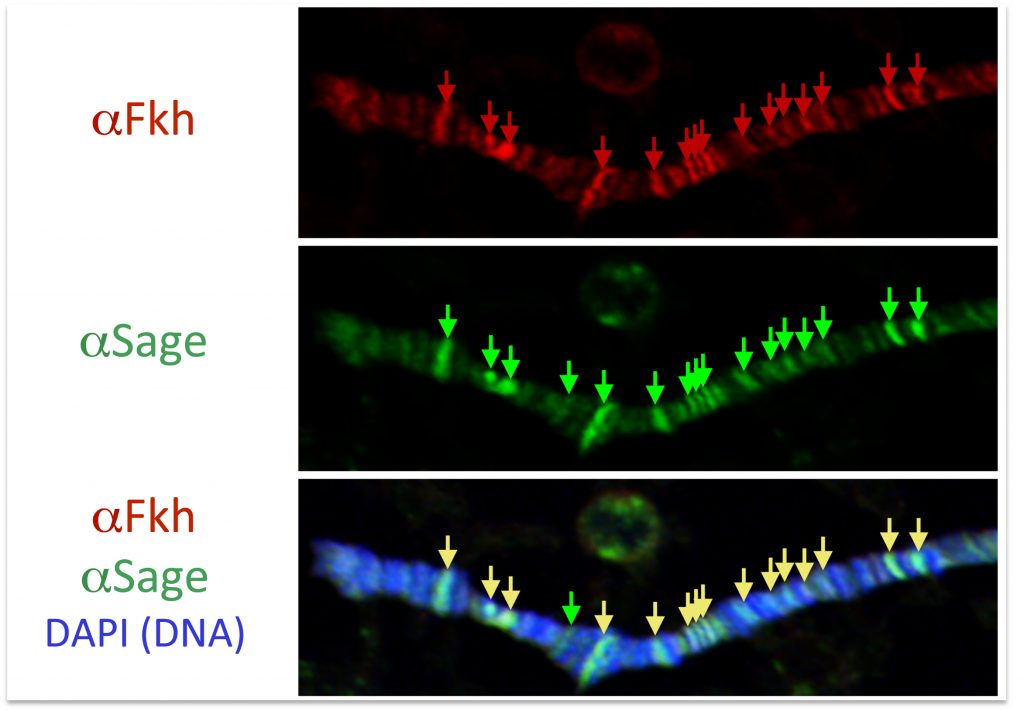
The Andrew Lab has demonstrated that Fork head (Fkh), the single member of the vertebrate FoxA family of transcription factors partners with Sage to activate transcription of the bZip transcription factor Senseless (Sens). In turn, Fkh, Sage and Sens activate high-level expression of SG-specific secreted cargo, transmembrane proteins and the SG-specific enzymes that modify these proteins as they traverse the secretory pathway. Low-resolution genome-wide chromatin binding assays (staining of SG polytene chromosomes with combinations of antisera directed against each transcription factor) suggest that the three proteins regulate their targets directly and in concert. High-resolution salivary gland-specific ChIP-Seq analysis (in collaboration with Matthew Slattery) is underway to uncover the mechanisms underlying the Fkh, Sage and Sens interactions with each other and with their shared downstream targets.
