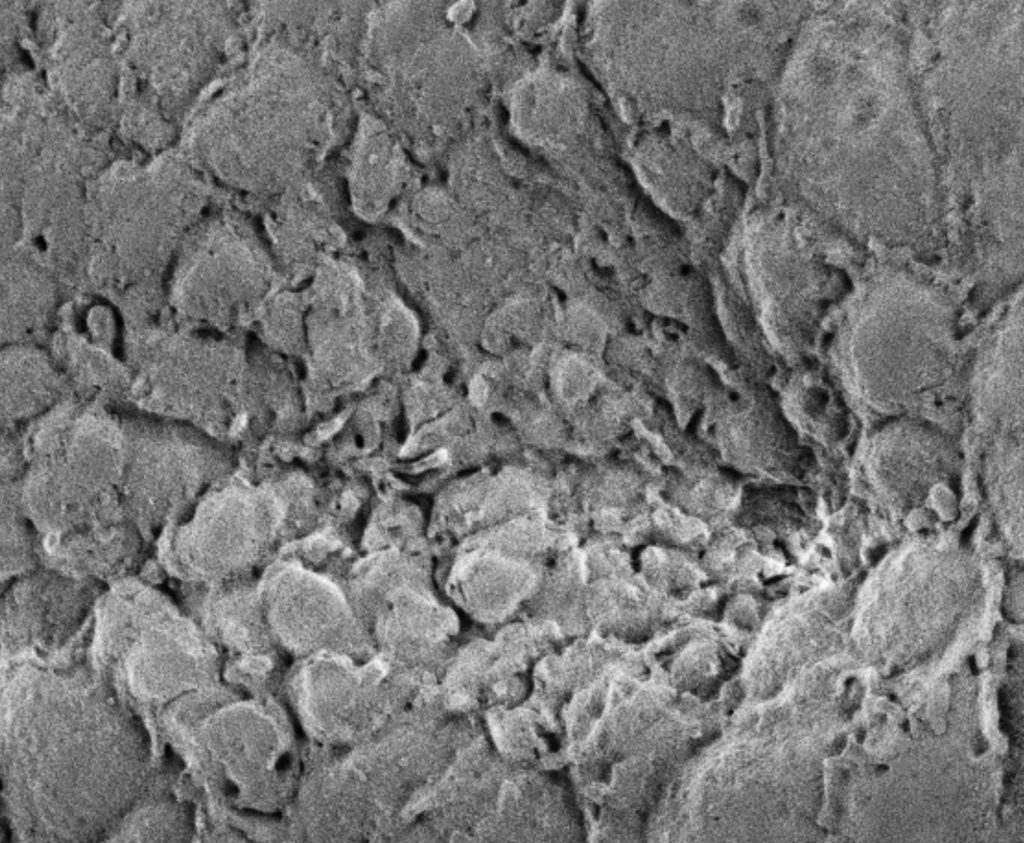
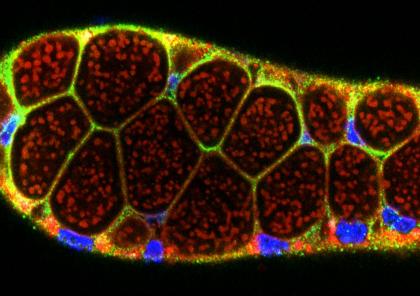
The Andrew Lab studies how organs are specified, form and specialize using the Drosophila salivary gland and trachea as model systems. We, and others, have identified the factors required to specify salivary gland and tracheal cell fates. Our lab has also characterized many of the earliest-acting tissue-specific transcription factors. These transcription factors maintain specialized cell fates, control morphogenesis, and activate tissue-specific gene expression patterns. In both the salivary gland and trachea, we have discovered many downstream targets of the early-expressed tissue-specific transcription factors. The functions of these target genes have revealed an unexpected parsing of cellular functions to different tissue-specific transcription factors and many of these functions are conserved among family members in higher eukaryotes. We continue to study the contributions of key downstream targets in both tissue specialization and the morphological transformation of tissue primordia into mature functional architectures. We are now also characterizing adult mosquito salivary glands from two human vector species as the first steps toward leveraging our findings on the Drosophila salivary gland to develop strategies to limit transmission of malaria and other mosquito-borne diseases.
The Andrew Lab uses a combination of genome-wide DNA binding and gene expression studies, classical and modern genetic and molecular biological approaches, as well as advanced microscopy and image analysis to uncover the gene networks governing organ development.
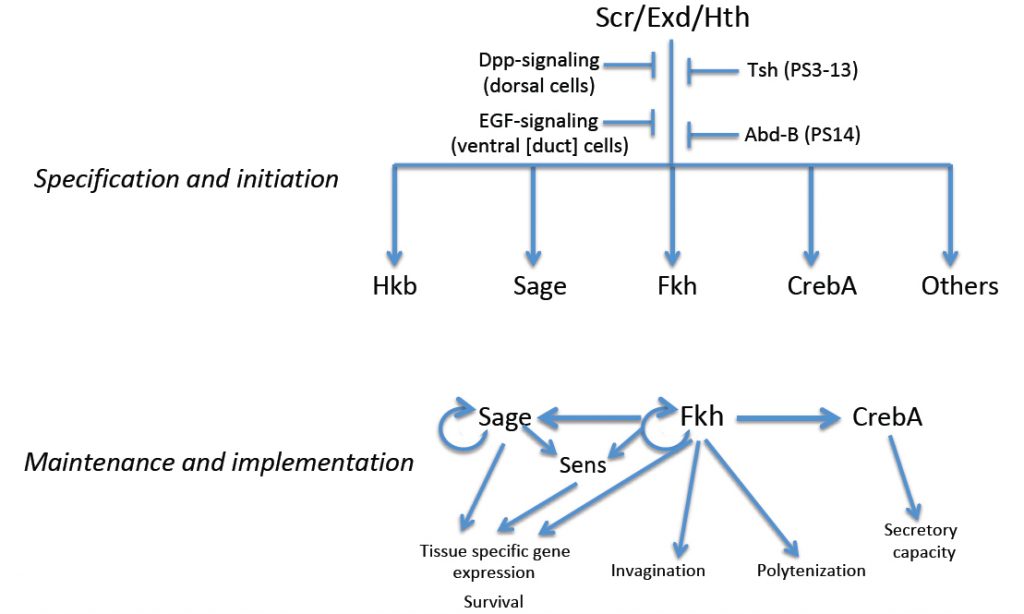
Drosophila salivary gland (SG) formation requires the Hox transcription factors, Sex combs reduced (Scr), Extradenticle (Exd) and Homothorax (Hth); in loss-of-function mutants for any of the corresponding genes, SGs fail to form. Moreover, ectopic expression of Scr, the one component of the complex with limited spatial expression, leads to the formation of additional SGs. Expression of Scr and hth and nuclear localization of Exd disappear, however, shortly after the SG initiates morphogenesis, suggesting that genes that specify the Drosophila SG cell fate are not involved directly in its terminal differentiation. Genes encoding several transcription factors are expressed in the early SG under the control of Scr, Exd and Hth, including fork head (fkh), which encodes a winged-helix transcription factor, CrebA, which encodes the Cyclic AMP Response Element Binding protein A, sage, which encodes an SG-specific bHLH protein, and huckebein (hkb), which encodes an Sp1/egr-like transcription factor. A subset of these early expressed SG transcription factors (Fkh, CrebA and Sage) maintain their own expression and/or the expression of each other. Thus, the early-expressed SG transcription factors both maintain and implement the SG cell fate decision, with Fkh playing a major role in this process.
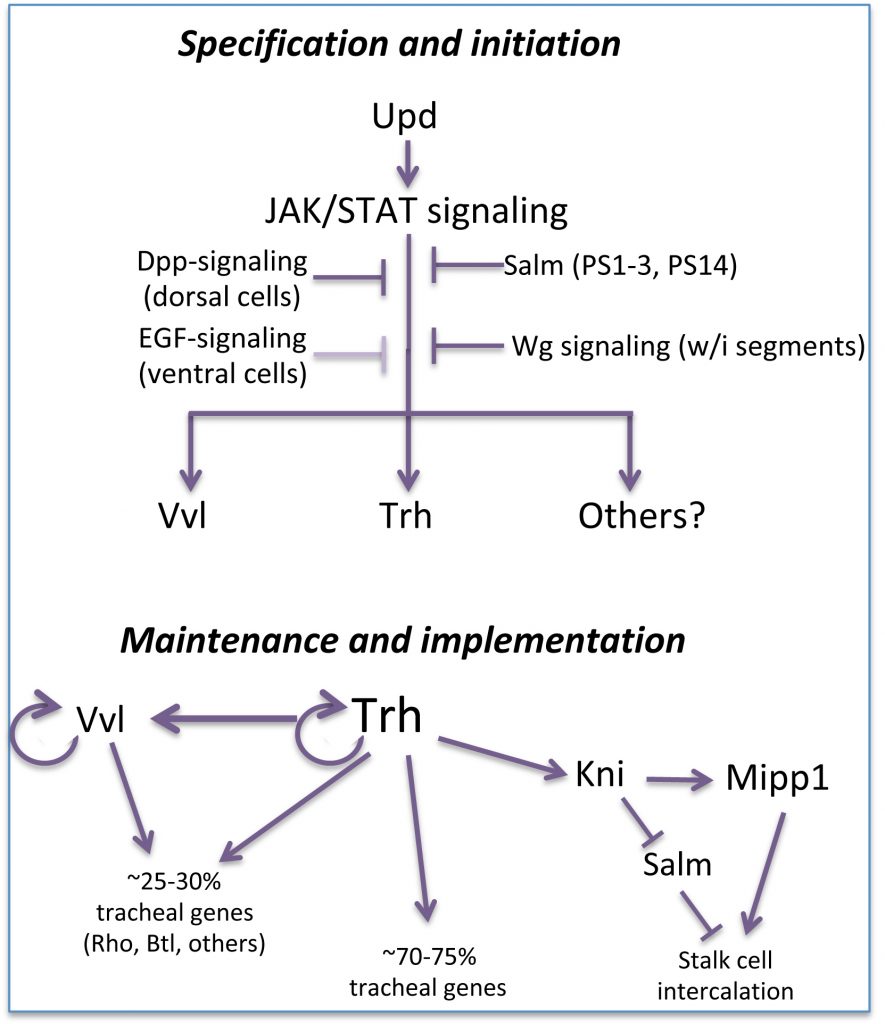
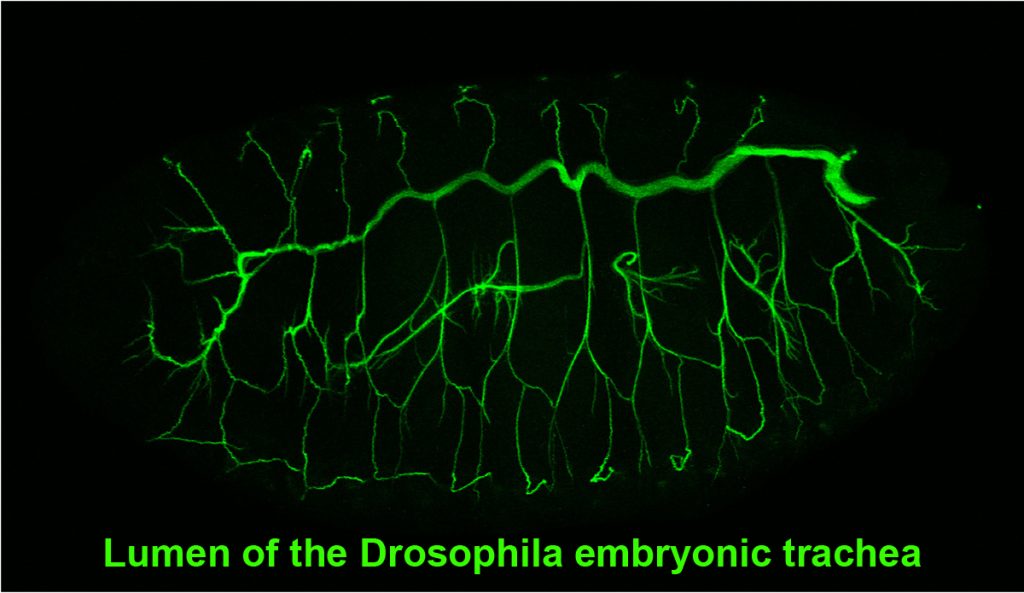
Drosophila embryonic trachea formation requires activation of the JAK/STAT signaling pathway by the Upd ligand. As with the SG, several factors, including the transcription factor Spalt as well as Wg-, Dpp- and EGF-signaling, limit trachea to only a subset of cells that experience JAK/STAT activation. Two major transcription factors are activated by the trachea-specifying genes: trachealess (trh) and ventral veinless (vvl). Both play major roles in maintaining and implementing the tracheal cell fate. Trh maintains its own expression and that of Vvl, and is required for expression of every known tracheal gene. Vvl is required for expression of an estimated 25-30% of tracheal genes. Thus, similar to the SG, multiple (at least two) transcription factors are required to both maintain and implement the tracheal cell fate decision, with Trh playing the major role in this process.
 The Andrew Lab has discovered that the CrebA/Creb3-like bZIP transcription factors are direct and major regulators of secretory capacity. Drosophila CrebA directly activates high-level expression of Secretory Pathway Component Genes (SPCGs) through a site we found to be conserved among the enhancers of CrebA-dependent genes. Moreover, ectopic expression of CrebA in multiple tissues is sufficient to activate high-level expression of every SPCG tested. Microarray analysis indicates that CrebA is required for full expression of ~400 genes including almost 200 already implicated in secretion. The secretory target genes include general machinery required for secretion in all cells as well as cell-type specific secreted cargo, such as the cuticle proteins and mucins. Phenotypic characterization of CrebA mutant SGs reveals a range of expected secretory defects, including reduced luminal secretory content and a decrease in the size and number of apical secretory vesicles, as well as unexpected changes in organelle distribution. In collaboration with Peter O’Hare’s group, we demonstrated that active forms of all five human CrebA orthologues: Creb3/Luman, Creb3L1/Oasis, Creb3L2/BBF2H7, Creb3L3/CrebH and Creb3L4/Creb4 can activate the Drosophila SPCGs when expressed in embryos. Active Creb3L1 also induces expression of multiple components of the secretory pathway when expressed in HeLa cells, a non-secretory cell type. These studies reveal an ancient and highly-conserved role for the CrebA/Creb3-like bZIP transcription factors in boosting secretory capacity. Current efforts are directed toward learning how CrebA interacts with target gene DNA in vivo (in collaboration with the research team of Matthew Slattery at the University of Minnesota, Duluth) and characterizing novel secretory pathway components identified based on their CrebA regulation.
The Andrew Lab has discovered that the CrebA/Creb3-like bZIP transcription factors are direct and major regulators of secretory capacity. Drosophila CrebA directly activates high-level expression of Secretory Pathway Component Genes (SPCGs) through a site we found to be conserved among the enhancers of CrebA-dependent genes. Moreover, ectopic expression of CrebA in multiple tissues is sufficient to activate high-level expression of every SPCG tested. Microarray analysis indicates that CrebA is required for full expression of ~400 genes including almost 200 already implicated in secretion. The secretory target genes include general machinery required for secretion in all cells as well as cell-type specific secreted cargo, such as the cuticle proteins and mucins. Phenotypic characterization of CrebA mutant SGs reveals a range of expected secretory defects, including reduced luminal secretory content and a decrease in the size and number of apical secretory vesicles, as well as unexpected changes in organelle distribution. In collaboration with Peter O’Hare’s group, we demonstrated that active forms of all five human CrebA orthologues: Creb3/Luman, Creb3L1/Oasis, Creb3L2/BBF2H7, Creb3L3/CrebH and Creb3L4/Creb4 can activate the Drosophila SPCGs when expressed in embryos. Active Creb3L1 also induces expression of multiple components of the secretory pathway when expressed in HeLa cells, a non-secretory cell type. These studies reveal an ancient and highly-conserved role for the CrebA/Creb3-like bZIP transcription factors in boosting secretory capacity. Current efforts are directed toward learning how CrebA interacts with target gene DNA in vivo (in collaboration with the research team of Matthew Slattery at the University of Minnesota, Duluth) and characterizing novel secretory pathway components identified based on their CrebA regulation.
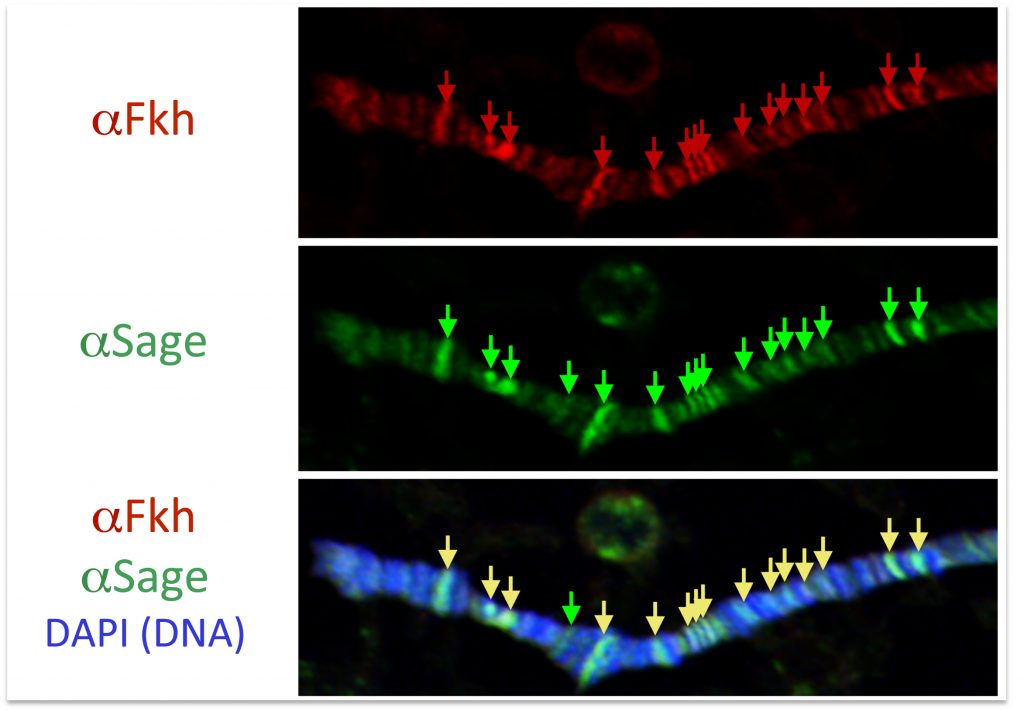
The Andrew Lab has demonstrated that Fork head (Fkh), the single member of the vertebrate FoxA family of transcription factors partners with Sage to activate transcription of the bZip transcription factor Senseless (Sens). In turn, Fkh, Sage and Sens activate high-level expression of SG-specific secreted cargo, transmembrane proteins and the SG-specific enzymes that modify these proteins as they traverse the secretory pathway. Low-resolution genome-wide chromatin binding assays (staining of SG polytene chromosomes with combinations of antisera directed against each transcription factor) suggest that the three proteins regulate their targets directly and in concert. High-resolution salivary gland-specific ChIP-Seq analysis (in collaboration with Matthew Slattery) is underway to uncover the mechanisms underlying the Fkh, Sage and Sens interactions with each other and with their shared downstream targets.
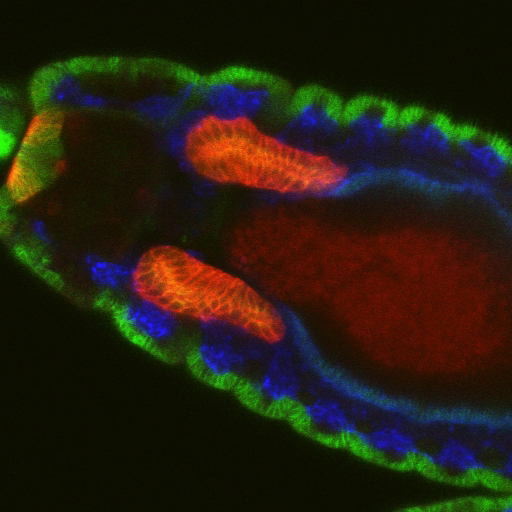
Salivary glands begin as two dimensional plates of ~144 polarized ectodermal cells each on the ventral surface of the embryo. Over a period of ~six hours, these two dimensional plates are converted into three-dimensional fully internalized and elongated tubes ~eight cells in circumference and ~8 cells long. Neither cell division nor cell death occurs once the SG has been specified, so the entire morphogenetic process must be driven by changes in cell shape, rearrangement, growth and collective migration. Fkh, Hkb and a BTB protein known as Ribbon (Rib) are each required for the formation of normal elongated secretory tubes. In fkh mutants, SGs completely fail to internalize and remain on the embryo surface. We have shown that Fkh regulates expression of the secreted ligand Folded Gastrulation (Fog) to coordinate apical constriction by controlling apical-medial pools of Rho Kinase and apical-medial Myosin activation. Activated apical-medial Myosin then drives apical constriction of SG cells as they internalize to form tubes through a budding-type mechanism. Although this coordinated apical constriction is required for the normal geometry of the nascent SG tubes, it is not essential for the primordia to internalize, indicating that other processes also contribute to SG tube formation. Indeed, we have shown that a supracellular myosin cable that surrounds the SG, first described by Katya Röper’s group, also contributes to internalization and that Fkh regulates expression of crumbs to drive this process. SG internalization also requires cells to change their neighbors while also remaining part of a completely polarized epithelium. We have recently identified two new Fkh-dependent target genes that contribute to SG cell rearrangement.
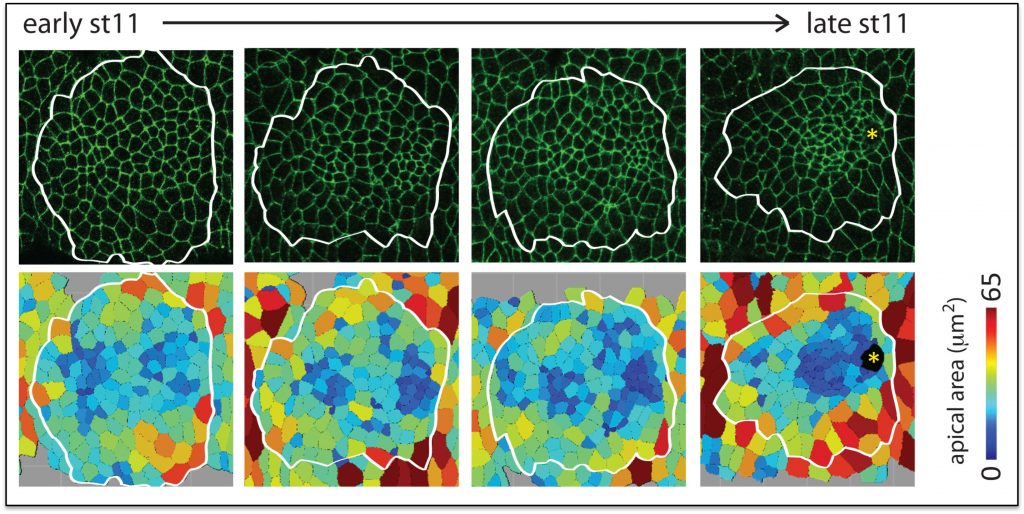
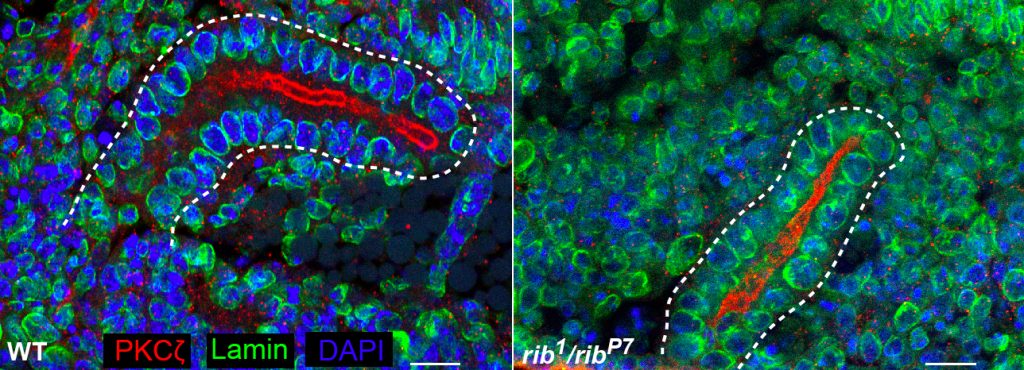
Hkb and Rib are required for full SG tube elongation. hkb mutant SGs internalize but fail to expand their apical surface, leading to the formation of small, puck-shaped SGs. Hkb controls apical membrane expansion through stabilization of Crumbs (Crb), an apical membrane determinant, and transcriptional upregulation Klarsicht, which mediates microtubule-dependent organelle transport. Rib mutant SGs also internalize but only ever achieve ~60% the length of WT SGs. Rib controls SG length both by regulating target genes that control apical membrane mechanics and by regulating target genes that mediate cell growth. Current efforts in the lab are directed toward finding and characterizing additional targets of Fkh, Hkb and Rib that contribute to building SGs of the correct shape, size and position in the embryo.
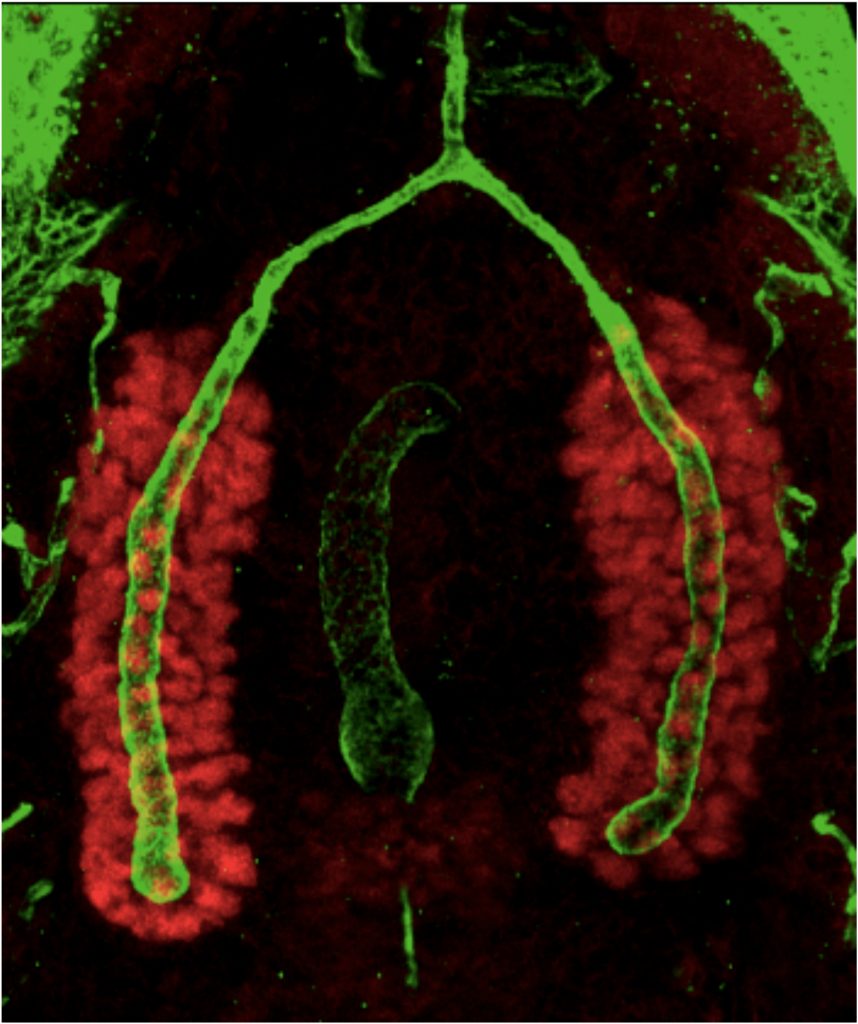
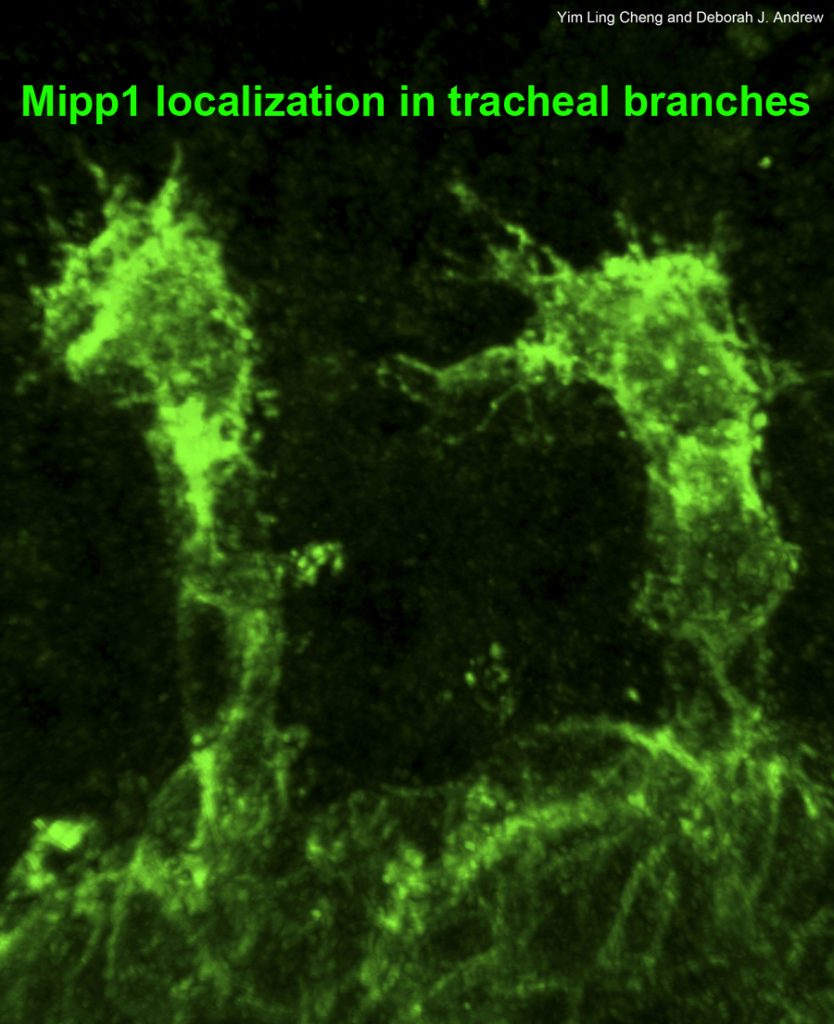
Tracheae begin as ten bilateral surface placodes in the early embryo. Shortly after the expression of Trh and Vvl, placode cells within each metamere (segment) invaginate to form internalized tracheal sacs of approximately 80 cells each. Tracheal cells subsequently undergo a series of stereotypical FGF-signaling dependent branching events. Cells at or near the ends of these branches eventually fuse with cells in neighboring tracheal segments to form an interconnected tubular network that delivers oxygen to every cell in the animal. Each tracheal segment has similar organization in terms of number of branches, number of cells within each branch, types of tubes each branch forms, and the migratory behavior of each branch. Branches are named for their positions and/or target tissues. Some branches, including the dorsal trunk (DT), elongate primarily, but not exclusively, by cell shape change. Other branches, including the dorsal branch (DB), elongate primarily by cell rearrangement. Among the Trh-Vvl shared targets is the FGF receptor gene breathless (btl), which contributes to tracheal invagination and is essential for the migration of all tracheal branches. Btl-expressing tracheal cells migrate toward target tissue sources of the FGF ligand Branchless (Bnl). Since Trh is the major transcription factor driving trachea formation, our efforts have been to identify and characterize additional downstream target genes. Among the Trh target genes we have discovered is Sano, an apically localized cytosolic protein that works with PCP genes to limit apical membrane expansion, and Mipp1, a highly conserved extracellular enzyme that converts inositol polyphosphates (IP6, IP5 and IP4) to IP3. Mipp1 is highly expressed in the leading cells of migrating tracheal branches where it functions extracellularly to mediate filopodia formation and confer migratory advantage. The characterization of additional Trh targets identified through genome-wide screens is underway.
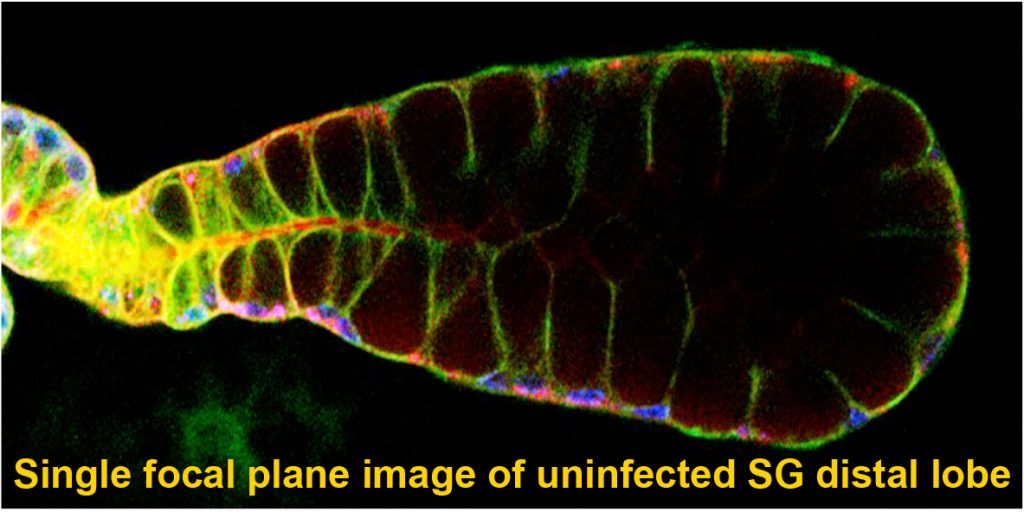
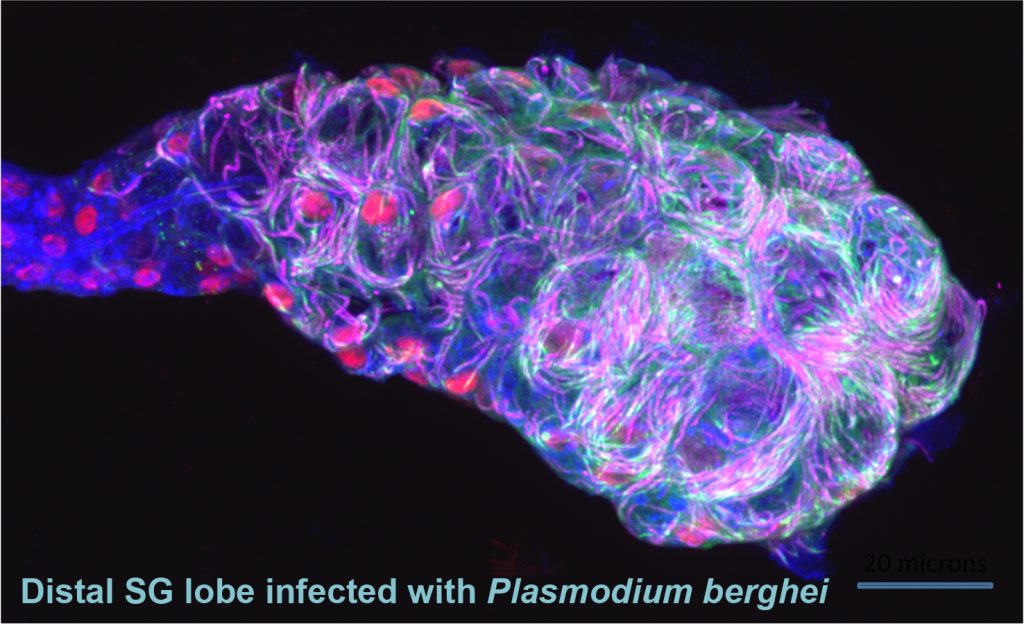
Mosquitoes and other insects are vectors for malaria, leishmaniasis, lyme disease, sleeping sickness, river blindness, Dengue fever and Zika – diseases that affect hundreds of millions of people and cause several million deaths every year. Most of these diseases require the disease-causing pathogen to traverse the insect salivary gland (SG) for transmission to humans. Despite the importance of the SG in the life cycle of insect-borne pathogens, almost nothing is known about the factors that drive the formation, function, and homeostasis of SGs in any insect outside of Drosophila. Our long-term goal is to develop and test strategies to block parasite transmission by intervening at the level of the mosquito vector, preventing transmission of disease by causing the timely death of the mosquito SG. Our studies leverage the considerable expertise of the Andrew lab regarding Drosophila SG biology to learn how mosquito SGs form and are maintained. Our initial goal is to determine if the mosquito versions of key transcription factors that function in Drosophila salivary gland specialization, survival and morphogenesis play the same critical roles in the adult mosquito SG. If so, our long-term goal is to target these genes in a tissue and time-specific manner as an additional strategy for limiting transmission of malaria. Importantly, this strategy should be generally applicable to preventing transmission of many insect borne human diseases.